Overview
The AcousticBrainz project makes available to the public the acoustic characteristics of music. The characteristics comprise of low-level spectral information and high level information about genres, moods, keys, scales etc. The high level information extraction is done using multiclass SVM models, which are trained using libsvm wrapped in a custom library called gaia written in C++. Gaia library is also used for classification and confusion matrix generation of recordings submitted by users in their own datasets. The machine learning capability of the system is right now limited due to it’s binding to C++ gaia library. With a view to extend it to use new machine learning algorithms and techniques such as deep learning, it is proposed to replace it with feature rich scikit-learn platform on python.
Personal Details:
Nickname: Nupur Baghel
IRC nick: nupurbaghel
Email: nupurbaghel@gmail.com
Github: nupurbaghel (Nupur Baghel) · GitHub
Existing gaia-based training process
Understanding existing system is very important to successfully complete this project. Therefore I have written below detailed description of current system.
Dataset Evaluation
Acousticbrainz portal has a facility for the users to submit their own recordings, having pre assigned musicbrainz-id to server using submission client. User then creates datasets , adds classes, attaches the submitted recordings to the classes and marks dataset for evaluation. The dataset is then picked up by multi threaded evaluation script running at server. Steps of execution are as below :
Project Classification :
The script reads low level data of recordings from the database table and dumps in yaml format to file system in a temporary directory. Below are the details of the files created :
-
project.yaml : Created using the template file. It stores :
A. filesystem location of dataset directory, results directory, filelist and groundtruth file.
B. Data preprocessing categories with details of transformation performed on various descriptors.
C. SVM classification parameters list i.e Type, kernel, C and gamma values to be used.
D. Evaluation method i.e. cross validation and number of folds currently 5. -
filelist.yaml : Mapping of recording id to low level info file path.
-
groundtruth.yaml : Mapping of dataset class to recording ids.
Model Training:
Preprocessing:
After creation of datasets directory and results directory, preprocessing is performed on the data. There are five categories of preprocessing: basic, low-level, nobands, normalized and gaussianized. Details of data transforms done during preprocessing are as below:
- Cleaner : Removes the data values which are either Infinite or NaN.
- FixLength : Removes descriptors which are not of fixed length within a single dataset.
- Removevl : Removes descriptors which have variable length across datasets like rhythm.beats_position.
- Remove : Removes the descriptors as listed in project file by pattern matching like ‘dmean’, ‘dvar’,‘.min’, '.max’, ‘*cov’. This is there in all five categories. “nobands” also performs removal of band related descriptors.
- Enumerate : Transforms string values such as tonal.chords_key, tonal.chords_scale, tonal.key_key, tonal.key_scale to numeric ones. It is done by sorting and using the ranks as values. This is also a part of all five categories.
- Normalize : Normalizes data to range [0,1] as it is essential for proper SVM classification.
- Gaussianize : Transforms data to form a perfect gaussian distribution. It is achieved by sorting the data, and giving them new values. The actual values are discarded and only ranks are kept.
Preprocessing steps are applied on original dataset and stored after each category of preprocessing as files with .db extension.
Training Details:
- Iterations are performed for each of the SVM parameter listed in project file. There are total 2 kernels, 9 C values, and 8 gamma values. i.e. total 298 = 144 iterations.
- Five fold cross validation evaluation is performed on the preprocessed data. The data is split into five parts, four parts are used for training and one part is used for testing. Each part is thus selected once for testing in five folds. Predictions are then performed on the dataset.
- Classification results are stored in results directory as “.result” and parameters as “.parameter” file. Name of the file is derived using a hash of training parameters.
Best model is chosen from the generated results files by calculating accuracy. Confusion matrix is derived as well. The training result thus comprises of parameters, accuracy, confusion matrix and history file location. The result is stored in database tables’s job result column. - The history file for the best model is saved as job_id.history.
- QString::fromUtf8 is used for saving history files.
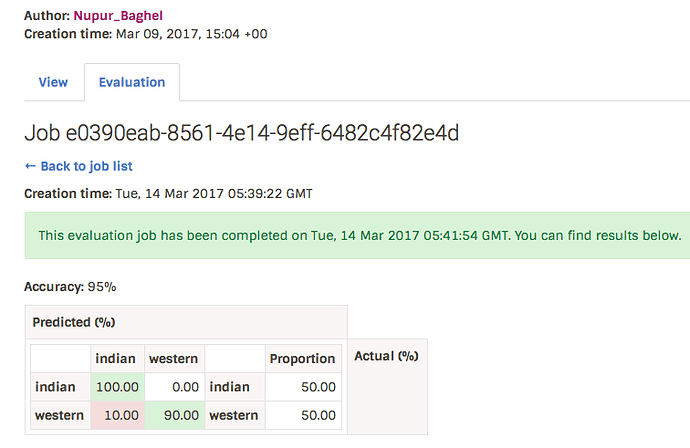
The confusion matrix is displayed on the website as below. The dataset had two classes named indian and western:
Highlevel Extractor
This is a another multithreaded task which keeps polling the database for recordings, whose high level data is awaiting extraction. The script uses a profile file template which comprises of details of all SVM model files , a total of 18 i.e.
danceability, gender, genre_dortmund, genre_electronic, genre_rosamerica, genre_tzanetakis, ismir04_rhythm, mood_acoustic, mood_aggressive, mood_electronic, mood_happy, mood_party, mood_relaxed, mood_sad, moods_mirex, timbre, tonal_atonal and voice_instrumental.
The script uses static binary extractor named streaming_extractor_music_svm.
The binary’s sha1 checksum is also written to the profile conf generated using the template file. Low level json data from database is dumped to a temporary file. The binary is then executed using the profile.conf file , low level data file and output file location as the input parameters. The script writes the high level data to the out file , which is written to the database when the job is over.
Highlevel Extractor Setup
Gaia, essentia and svm models are required to be downloaded from respective git repositories, compiled and installed. Extractor setup script lists the other dependencies to be installed as well.
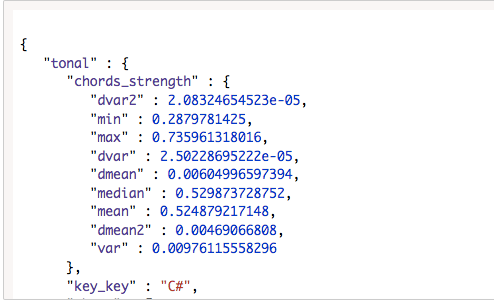
Low level data :
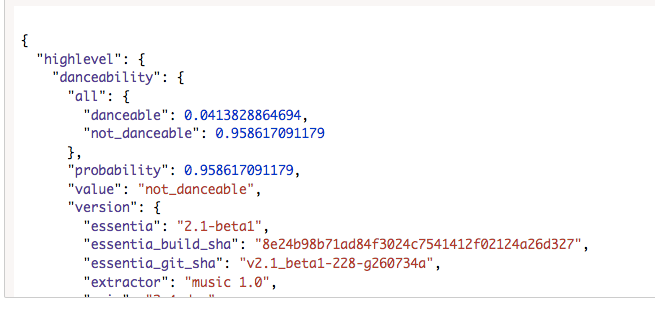
High level data :
Proposed Scikit-Learn Infrastructure
This project is about replacing existing gaia based machine learning with scikit-learn based system. Being a high priority project the task has already been started. It is proposed to complete it and enhance further to the extent possible. The project can be divided in four subparts:
- Development of stand alone model training system
- Comparison with gaia results and applying corrections
- Development of dataset evaluator to work with scikit trainer
- Development of high level extractor using scikit trainer
Task already completed from subpart 1 :
- Creation of project yaml file from the template, filelist and groundtruth file creation.
- Remove and Enumerate transform functions.
- Flattening multi level dictionary to single level for applying to Scikit SVM classifier.
- Iterating over preprocessing categories and SVM parameter combinations.
- Invoking Scikit SVM classifier
- Model serialization using pickle.
Proposed Development Details :
Milestone 1 : Development of stand alone model training system :
This part will be divided into modules as below:
Preclassification Module:
This will generate project yaml file, filelist and groundtruth file.
Transform Module:
This module will have a base class with abstract interface describing purpose of the transformation, which will be inherited by actual transform classes as listed below:
Cleaner:
To remove descriptors that are either constant values or contain NaN or Inf values. This may use numpy.sum() function.
RemoveVariableLength:
To remove descriptors having variable length across different low level files. The input will be a dictionary and output will be a subset after performing removal.
Enumerate:
To convert string values to numeric, as the input data to SVM classifier must be numeric.
Select:
To have the desired descriptors and exclude the ones not required or each type of preprocessing. This class will return a flattened dictionary having just one level of keys and values.
Normalize:
To normalize data to [0, 1] using Scikit’s sklearn.preprocessing.minmax_scale
Gaussianize:
To apply gaussian transformation on data. This will be coded in python on similar lines as existing gaia code in file gaia/src/algorithms/gaussianize.cpp.
Training Module:
This module will be performing the model trainer by using the functions below :
load_and_initial_transform:
Loads project filelist, then calls transforms Cleaner, RemoveVariableLength, Select and Enumerate. Returns a dictionary of {filekey: filtereddata, …}
enumerate_combinations
Iterates over all preprocessing categories and list of kernel, C and gamma values.
cross_validate
Use built in cross_val_predict from sklearn.model_selection: cross_val_predict(clf, X, cv=5). Input parameters are the fitted classifier and the training dataset. cross_val_score will be used to calculate cross validation accuracies by applying mean function on scores. Returns the predicted class matrix and the the accuracy.
save_result:
This will store the predicted result matrix in a result file similar to gaia.
save_param:
This will store the SVM parameters used to train the model in a parameter file.
find_best_model:
Goes through all result files. Finds the best model by calculating accuracy and returns result.
save_model
Stores serialised model.
train_model:
This is the function called from main evaluate script. It calls functions above in the sequence: load_and_initial_transform , enumerate_combinations , normalize and gaussianize transforms as needed by the preprocessing category, map_class to convert class names to numbers. Creates SVM classifier : clf = SVC(kernel=k, gamma=2g, C=2c) and clf.fit(data,classes) and calls cross_validate, save_result , save_param, find_best_model and then save_model.
Serialisation Module
The details of model training will have to be stored so that it can be applied later on the test data. We can use Python’s Pickle module for this purpose .This module will be used spcifically for storing :
The trained SVM model as recommended by scikit-learn for model persistence.
To store a mapping between the numerical class identifiers which scikit-learn uses and the text class names used in AcousticBrainz datasets. Because input data needs to be transformed in the same way when predicting an unknown item as it was when the model was trained.
To store mapping between the text and number from the Enumerate module of transform section.
Milestone 2 : Comparison with gaia results and applying corrections
In order to verify that the implementation of the scikit-learn training system is correct we need to evaluate it against the existing Gaia system. I will do this by developing a script which takes a single dataset and generates two models, one using Gaia and the other using scikit-learn.
An evaluation script will check that the same transforms are made, and that the same parameters are used in training the model. I will need to check the preprocessing category applied, kernel, gamma and C values used at minimum. There could be some parameters as well , which I am not aware at this stage.
The script will check that the cross-validation accuracy between the two systems is the same, and that a random unknown low level file results in the same prediction with both systems.
Scikit trainer will be corrected according to the results of evaluation script.
Milestone 3 : Development of dataset evaluator to work with scikit trainer
The script will pick up jobs pending evaluation from the database and pass to the trainer one by one . The status of the job will be set to “running”. After the evaluation is complete, the results i.e. project_path, svm parameters used, accuracy, confusion_matrix , the location of trained model file and may be few more details, will be written back to the database. Job status will be set to “complete”.
Milestone 4 : Development of high level extractor using scikit trainer
Highlevel extractor needs the pre trained models to perform classification on the 18 attributes listed above. For development of models using scikit trainer, I will need access to the data used earlier to generate those models. As this data is not available openly, I will request access to data for a couple of datasets i.e. “danceability” and “genre_dortmund”. The models will then be generated using the new dataset evaluator.
I will then develop the extractor script for the new trainer on the same lines as the current one. Though I need to perform more detailed study of current static binary used to extract high level data i.e. datastreaming_extractor_music_svm, at broader level this script calls the trained model, performs testing and produces results. For example if the test is being performed for “danceability”, the script will tell how much positive the result is towards danceability and then the final result.
The high level data thus comprises the results of testing using all the models. Also it comprises the version of software read from the configuration file, the git sha value etc.
To make sure that the extractor is working correctly, I will develop a script which calls both gaia extractor and the scikit extractor on the same dataset and produces results in the form of a comparison table. Corrections will be applied based on the results.
The final extractor script will be able to produce the high level data file similar to the existing one and also write the high level data to the database.
Milestone 5 : Offline job running client (in case development of above 4 milestones is completed before time):
As the work has already been started on my main project, If it so happens that a big chunk of jobs listed above gets completed before start of Soc period, I propose to complete the offline job running client using the scikit trainer. It was a partially completed project done in GSoc 2016 using Gaia classifier. It was basically meant to perform the dataset evaluation locally in user’s own system and submit the results back to server. Below components will be used as it is :
-
Submitting jobs for evaluation: Dataset evaluation page on acousticbrainz portal has a button to submit jobs for evaluations locally. The button is managed by config parameter FEATURE_EVAL_LOCATION , which is currently turned off.
-
Client side code to download jobs: acousticbrainz-dataset-runner/dataset_runner/run_jobs.py at master · metabrainz/acousticbrainz-dataset-runner · GitHub
-
API to be called by client to download the jobs: acousticbrainz-server/webserver/views/api/v1/dataset_eval.py at master · metabrainz/acousticbrainz-server · GitHub.
-
Script to generate low level data file for recordings : available in scikit model trainer under development.
Here are the things i will be developing new :
-
Write script similar to current evaluate.py, this script will be called after downloading the jobs as in step 3 above. The jobs will be submitted to the new scikit model trainer. The results will be stored in a file locally with dataset id, job_id and user_id.
-
API script to submit the results to server via POST request. The script will check, if the job belongs to the user and is pending evaluation at user’s local system. It will also perform a structure check on the submitted result. It will then update the database to store the result for that dataset and also change the status of the job to “Done”.
-
User will have to clone the repository containing the bundle of software. Two scripts will be available to execute , one for starting the job and the other for submitting the results back to the server.
Proposed Project Timeline:
Community Bonding (May 5 - May 29)
Pre-complete tasks from Phase 1 as far as possible. Study high level extractor. Keep track with the ongoing development.
Phase 1 (May 30 - June 25) : Milestone 1.
Week 1: Complete transform module.
Week 2: Complete training module functions other than train_model.
Week 3: Complete train_model.
Week 4: Test and bug fix development so far. PHASE 1 evaluation takes place.
Phase 2 (June 26 - July 30) : Milestone 2 and 3.
Week 5: Develop script to call gaia and scikit classifier and the evaluation script.
Week 6: Testing and comparison with gaia results and corrections
Week 7: Develop new dataset evaluator.
Week 8: Testing and bug fixing dataset evaluator. PHASE 2 evaluation takes place.
Phase 3 (August 1 - August 29) : Milestone 4
Week 9: Develop extractor script for the new trainer.
Week 10: Develop script to invoke gaia and scikit extractor and the comparison page.
Week 11: Perform testing and applying corrections.
Week 12: Documentation , changes in the installation manual of accousticbrainz app.
Week 13: Final submission.
Milestone 5 will be done as a part of phase 3, incase work of previous milestones gets completed before time , because of early start of this project.
Details about myself:
-
Tell us about the computer(s) you have available for working on your SoC project!
I will be working on a mac-book air. -
When did you first start programming?
I started programming with c++ during my higher schooling , four years back in 2013.
Did a bit of html in the year prior to that. -
What type of music do you listen to? (Please list a series of MBIDs as examples.)
I love listening bollywood as well as english songs. Say
e387ac45-4be6-4b4f-a957-ed0fe0afa3ca, d4b11f49-755a-4ace- b22d-3c3a5112f639, e0b5fffd-b278-4f0c-88f7-3e98519699c7, 9086b742-358b-4f73-9a14-84cb1a9ce4ce . -
What aspects of the project you’re applying for (e.g. MusicBrainz, AcousticBrainz etc. ) interest you the most.
With this project i will get an opportunity to implement the sci-kit library and its extensive ML algorithms to process low-level and high-level data which will make the application easier to extend with techniques like big learning. -
Have you ever used MusicBrainz to tag your files?
Yeah, when i tried to submit music files to my local server, I used Picard to generate mbids. -
Have you contributed to other Open Source projects? If so, which projects and can we see some of your code?
No, Metabrainz is my first Open Source project, I have started contributing to it already. One of my pull requests has been merged and three others are under progress. -
If you have not contributed to open source projects, do you have other code we can look at?
Yes, I have participated in contests and worked at the web-team of my college. I made a small blog application using flask and deployed it to heroku.
GitHub - nupurbaghel/nupur-blog: This is a small blog app, through which users can make new blogs and posts., https://nupblog.herokuapp.com -
What sorts of programming projects have you done on your own time?
I have done mini-projects like building chatbots for downloading newsfeeds , images and music. I am also working on an ML project titled “Pattern recognition algorithm to identify marine animals” under Smart India Hackathon, which deals with image detection and processing using opencv. -
How much time do you have available, and how would you plan to use it?
I will be spending 8 to 10 hours on a daily basis completely during summers and 6 hours or more during my college days. -
Do you plan to have a job or study during the summer in conjunction with Summer of Code?
No, I plan to work fully for the Soc project during summers.
I am looking forward to get valuable feedback from the members of the community.